
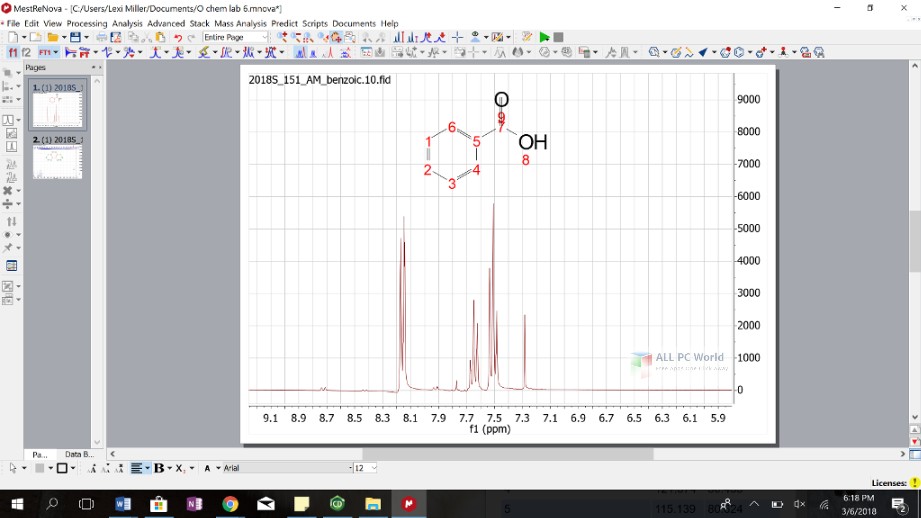
Reference the 1 H spectrum using TMS or the residual solvent (CHCl 3, d 5 -DMSO, etc.) peak, and reference the 13 C spectrum using the solvent peak (CDCl 3, d 6 -DMSO, etc.). For a homonuclear (COSY, ROESY, NOESY, TOCSY) spectrum you will want a 1 H spectrum in both dimensions.Ģ Very Important: First, set the reference for the 1D spectra before using them as traces. For a heteronuclear inverse (HSQC or HMBC) spectrum, you will want a 1 H spectrum in the F 2 dimension and a 13 C spectrum for the F 1 dimension.
MestReNova calls these 1D spectra Traces. You will want to have a 1D spectrum corresponding to the F 2 dimension nucleus displayed along the top of the 2D rectangle, and another corresponding to the F 1 dimension nucleus displayed along the left side. Click on 2D (at the left) and select Red-Blue for Palette, Contour for Plotting method, 10 for Number of Positive Contours, 10 for Number of Negative Contours, and for Scaling and 8.0 for Line Width. Right-click on your 2D spectrum and select Properties (near the bottom). For consistency, always use red for positive intensities and blue for negative. It will take a few seconds as it re-processes the 2D data. Click OK on the Processing Template window. Click on the next button ( ) down, next to Spectrum Size, and set to Click OK. Click on the first button ( ) under the Apodization heading, and set the F 1 window function as follows: COSY: Sine Bell, 0.00 Deg HSQC or TOCSY: Sine Bell, Deg HMBC (magnitude mode): Sine Square, 0.00 Deg ROESY or NOESY: Sine Bell, Deg Be sure to turn off (uncheck) any other Apodization settings besides the one you want (Sine Bell or Sine Square). Make sure that the Truncate box is NOT checked. Click on the next button down, next to Spectrum Size, and set to Click OK. Set the window function in F 2 as follows: COSY: Sine Bell, 0.00 Deg HSQC or TOCSY: Sine Bell, Deg HMBC, ROESY or NOESY: Sine Bell, Deg (45.00 Deg if S/N is good) Be sure to turn off (uncheck) any other Apodization settings besides Sine Bell. With the f2 tab highlighted, Click on the button to the right and just below Apodization. Click on the drop-down menu Processing and select Processing Template. These parameters control the peak shape and resolution in both dimensions, and different values are required depending on the type of 2D spectrum.
For Varian data just drag the filename.fid folder into the MestReNova window. Drag this folder into the main window of MestReNova. On your PC or Mac you can rename the Bruker Experiment Number folder (1, 2, 3 ) to HSQC, COSY, HMBC, ROESY, etc. 1 Processing 2D NMR Data with MestReNova 1.


 0 kommentar(er)
0 kommentar(er)
